Career Interests
- Genome editing for hematological diseases
- Base editing, Prime editing
- Genome modification in human stem cells including hematopoietic stem cells
Education
- Indo-US GETin fellow, Innovative Genomic Institute (IGI), UC Berkeley, California, USA.
- Post-Doctoral Research Fellow - 2007-2015 St. Jude Children’s Research Hospital, Memphis, TN, USA.
- PhD in Molecular Medicine (Breast Cancer Biology) - 2004- 2007 Liggins Institute, University of Auckland, Auckland, New Zealand.
- MSc in Medical Biochemistry - 1997-2000 JIPMER, Pondicherry, India.
Awards
- Ben Barres Spotlight Award from eLife (2022)
- Indo-US Genome Engineering/Editing Technology Initiative (GETin) fellowship.
- Kelliher Trust Award for Research Innovation.
- International Doctoral Scholarship, University of Auckland, New Zealand.
- Maurice and Paykel Trust fellowship.
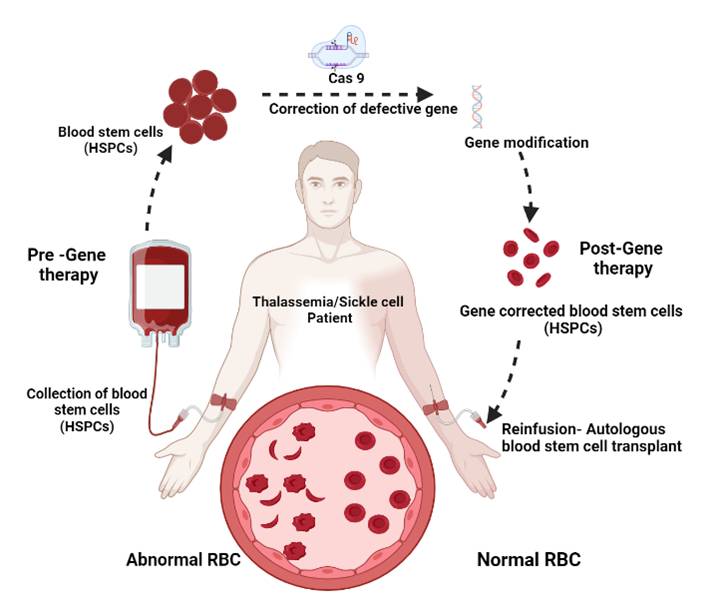
My laboratory is interested in developing innovative therapeutic genome editing tools to treat patients with inherited hematological disorders such as sickle cell diseases, thalassemia and Hemophilia A&B. We are planning to directly manipulate the genome of human hematopoietic stem cells (HSCs) in a sequence specific manner using targeted genome engineering platform based on CRISPR/CAS9 system for the treatment of inherited hematological disorders. We are further working on the delivery platforms to increase the genome editing efficiency and reduce the off target effects. We anticipate that our strategy will overcome the therapeutic barriers that currently limit the clinical applications.
1.Preclinical genome editing approach for the treatment of beta-globin disorders
Hemoglobinopathies such as sickle cell disease (SCD) and β-thalassemia are the most common genetic disorders in India. Elevation of fetal haemoglobin in SCD and β-thalassemia patients offers greater therapeutic advantage by ameliorating clinical symptoms. We utilize targeted genome engineering platform based on CRISPR/CAS9 system to reactivate gamma globin by editing the two potent gamma globin regulatory regions in hematopoietic stem cells for the treatment of SCD and thalassemia.
(I). Non deletional HPFH
Non deletional HPFH mutations are caused by point mutations in the HBG1 and HBG2 gene promoters. These point mutations greatly increase the production of Aγ-globin or Gγ-globin chain, which is normally silenced in humans after birth. In this context, we are using CRISPR- base editing approach to recreate the naturally occurring HPFH-associated mutation in human blood progenitors for the therapeutic induction of fetal hemoglobin level. In addition, we screened the proximal promoter of human HBG genes using adenine and cytosine base editors to identify key nucleotide point mutations that could potentially lead to elevated levels of fetal globin. Through systematic tiling across the HBG proximal promoter, we identified multiple novel target sites that resulted in a significant increase in fetal globin levels. This work shed light on so far unknown regulatory elements within the HBG promoter that normally mediates fetal globin silencing and identified additional targets for therapeutic upregulation of fetal hemoglobin
(II) Fetal globin repressors
Several transcription factors including BCL11A play a vital role through modulation of fetal globin level. However, BCL11A is dispensable in non-erythroid functions such as for normal lymphoid and neural development. Functional mapping of the Bcl11A enhancer identified the minimal critical sequence that is specific for erythroid specific BCL11A expression. For the targeted editing of fetal globin repressors, we are editing of erythroid specific BCL11A enhancer in mobilized hematopoietic stem and progenitor cells (HSPCs) from sickle cell disease, beta thalassemia and normal donor using the CRISPR/cas9 system.
2.Genome engineering hematopoietic stem cells for the treatment of Hemophilia A
Hemophilia A (HA) is an X-linked monogenic congenital bleeding disorder due defective FVIII in the bloodstream. Currently available therapeutic options include clotting factor replacement either with functional recombinant protein or with delivering FVIII gene in viral vectors. However, they are limited with immune tolerance induction and preexisting antibodies respectively. Even a small increase in FVIII levels can significantly ameliorate disease phenotype and patient’s quality of life, making this disease an optimal target for CRISPR/cas9 based genome editing system. To improve the current approaches of gene therapy for Hemophilia A, we are working on a novel ex vivo gene therapy approach for targeted integration of FVIII in hematopoietic stem cells for the treatment Hemophilia A.
- George A, Sadanandan P, Ravi NS, Vaishnavi B, Marepally S, Thangavel S, Velayudhan SR, Srivastava A, Mohankumar KM. Editing of homologous globin genes by nickase-deficient base editor mitigates large intergenic deletions in HSPCs. Molecular Therapy Nucleic Acids. 2024 Sep 30;35(4):102347. doi:10.1016/j.omtn.2024.102347. eCollection 2024 Dec 10. https://doi.org/10.1016/j.omtn.2024.102347
- Arjunan P, Mahalingam G, Sankar P, Kathirvelu D, Suresh S, Rani S, Mohankumar KM, Thangavel S, Marepally S .Base-modified factor VIII mRNA delivery with galactosylated lipid nanoparticles as a protein replacement therapy for haemophilia A. Biomater Science. 2024 Sep 25;12(19):5052-5062 doi:10.1039/d4bm00909f.PMID: 39210734 https://doi.org/10.1039/d4bm00909f
- Kirti Prasad, Nivedhitha Devaraju, Anila George, Nithin Sam Ravi, Joshua Paul P, Gokulnath Mahalingam, Vignesh Rajendiran, Lokesh Panigrahi, Vigneshwaran Venkatesan, Kartik Lakhotiya, Yogapriya Periyasami, Aswin Anand Pai, Yukio Nakamura, Ryo Kurita, Poonkuzhali Balasubramanian, Saravanabhavan Thangavel, Shaji R Velayudhan, Srujan Marepally, Alok Srivastava, Mohankumar KM.(2024) Precise correction of a spectrum of β-thalassaemic mutations in the coding and non-coding regions by base editors. Molecular Therapy - Nucleic Acids. (Online). https://doi.org/10.1016/j.omtn.2024.102205
- Vignesh Rajendiran, Nivedhitha Devaraju, Nithin Sam Ravi, Lokesh Panigrahi, Joshua Paul, Chandrasekar Gopalakrishnan, Stacia Wyman, Keerthiga Ariudainambi, Gokulnath Mahalingam, Yogapriya Periyasami, Kirti Prasad, Anila George, Dhiyaneshwaran Sukumaran, Sandhiya Gopinathan, Aswin Anand Pai, Yukio Nakamura, Poonkuzhali Balasubramanian, Rajasekaran Ramalingam, Saravanabhavan Thangavel, Shaji R Velayudhan, Jacon E Corn, Merlin Crossley, Srujan Marepally, Alok Srivastava, Mohankumar KM. (2024) Base editing of key residues in the BCL11A-XL-specific zinc finger domains de-represses fetal globin expression. Molecular Therapy. Mar 6;32(3):663-677. doi: 10.1016/j.ymthe.2024.01.023. https://doi.org/10.1016/j.ymthe.2024.01.023
- Mahalingam, G, Rachamalla, H. K, Arjunan, P, Karuppusamy, K. V, Periyasami, Y., Mohan, A, Subramaniyam, K, M, S., Rajendran, V, Moorthy, M, Varghese, G. M, Mohankumar KM, Thangavel, S, Srivastava, A, & Marepally, S. (2024) SMART-lipid nanoparticles enabled mRNA vaccine elicits cross-reactive humoral responses against the omicron sub-variants. Molecular Therapy. 32(5), 1284–1297. https://doi.org/10.1016/j.ymthe.2024.02.028
- Nandy K, Babu D, Rani S, Joshi G, Ijee S, George A, Palani D, Premkumar C, Rajesh P, Vijayanand S, David E Mohankumar KM, Shaji R Velayudhan. (2023) Efficient gene editing in induced pluripotent stem cells enabled by an inducible adenine base editor with tunable expression. Scientific Reports.11;13(1):21953. https://doi.org/10.1038/s41598-023-42174-2
- Nithin Sam Ravi, Anila George, Mohankumar KM. (2023), Arrayed gRNA screening by base editors in mammalian cell lines using lentiviral system, Star Protocol. 4(4):102668. https://doi.org/10.1016/j.xpro.2023.102668
- Mahalingam G, Arjunan P, Periyasami Y, Dhyani AK, Devaraju N, Rajendiran V, Christopher AC, Kt RD, Dhanasingh I, Thangavel S, Mohankumar KM, Moorthy M, Srivastava A, Marepally S. (2023) Correlating the differences in the receptor binding domain of SARS-CoV-2 spike variants on their interactions with human ACE2 receptor. Scientific Reports. 30;13(1):8743. https://doi.org/10.1038/s41598-023-35070-2
- George A, Ravi NS, Prasad K, Panigrahi L, Koikkara S, Rajendiran V, Devaraju N, Paul J, Pai AA, Nakamura Y, Kurita R, Balasubramanian P, Thangavel S, Marepally S, Velayudhan SR, Srivastava A and Mohankumar KM. (2022) Efficient and error-free correction of sickle mutation in human erythroid cells using prime editor-2. Frontiers in Genome Editing. 20:4:1085111. https://doi.org/10.3389/fgeed.2022.1085111
- Lohchania B, Christopher AC, Arjunan P, Mahalingam G, Kathirvelu D, Prasannan A, Venkatesan V, Taneja P, Mohankumar KM , Thangavel S, Marepally S.(2023) Diosgenin enhances liposome-enabled nucleic acid delivery and CRISPR/Cas9-mediated gene editing by modulating endocytic pathways. Frontiers in Bioengineering and Biotechnology 9;10:1031049. https://doi.org/10.3389/fbioe.2022.1031049
- David, E., Nakamura, Y., Balasubramanian, P., Srivastava, A., Thangavel, S., Mohankumar, K.M., Velayudhan, S.R., 2022. Erythroid lineage-specific lentiviral RNAi vectors suitable for molecular functional studies and therapeutic applications. Scientific Reports. 12, 14033. https://doi.org/10.1038/s41598-022-13783-0
- Mahalingam, G., Rachamalla, H.K., Arjunan, P., Periyasami, Y., M, S., Thangavel, S., Mohankumar, K.M., Moorthy, M., Velayudhan, S.R., Srivastava, A., Marepally, S., 2022. Optimization of SARS-CoV-2 Pseudovirion Production in Lentivirus Backbone with a Novel Liposomal System. Front. Pharmacol. 13, 840727. https://doi.org/10.3389/fphar.2022.840727
- Venkatesan V, Christopher AC, Rhiel M, Azhagiri MKK, Babu P, Walavalkar K, Saravanan B, Andrieux G, Rangaraj S, Srinivasan S, Karuppusamy KV, Jacob A, Bagchi A, Pai AA, Nakamura Y, Kurita R, Balasubramanian P, Pai R, Marepally SK, Mohankumar KM, Velayudhan SR, Boerries M, Notani D, Cathomen T, Srivastava A, Thangavel S. Editing the core region in HPFH deletions alters fetal and adult globin expression for treatment of β-hemoglobinopathies. Mol Ther Nucleic Acids. 2023 Apr 26;32:671-688. doi: 10.1016/j.omtn.2023.04.024. https://doi.org/10.1016/j.omtn.2023.04.024
- Ravi NS, Wienert B, Wyman SK, Bell HW, George A, Mahalingam G, Vu JT, Prasad K, Bandlamudi BP, Devaraju N, Rajendiran V, Syedbasha N, Pai AA, Nakamura Y, Kurita R, Narayanasamy M, Balasubramanian P, Thangavel S, Marepally S, Velayudhan SR, Srivastava A, DeWitt MA, Crossley M, Corn JE, Mohankumar KM (2022) Identification of novel HPFH-like mutations by CRISPR base editing that elevates the expression of fetal hemoglobin. eLife. 2022 Feb 11;11:e65421. doi: 10.7554/eLife.65421. https://doi.org/10.7554/eLife.65421
- Karuppusamy K, Demosthenes J, Venkatesan V, Christopher A, Babu P, Azhagiri M, Jacob A, Ramalingam V, Rangaraj S, Mohankumar K.M, Marepally S, Varghese G, Srivastava A, Kannangai R and Thangavel S. The CCR5 gene-edited CD34+CD90+ Hematopoietic stem cell population serves as an optimal graft source for HIV gene therapy. Frontiers in Immunology. 14:13:792684. https://doi.org/10.3389/fimmu.2022.792684
- Christopher AC, Venkatesan V, Karuppusamy KV, Srinivasan S, Babu P, Azhagiri MKK, Chambayil K, Bagchi A, Rajendiran V, Ravi NS, Kumar S, Marepally SK, Mohankumar KM, Srivastava A, Velayudhan SR, Thangavel S (2022) Preferential Expansion of human CD34 + CD133 + CD90 + Hematopoietic Stem Cells Enhances Gene-Modified Cell Frequency for Gene Therapy. Human Gene Therapy. 2022 Feb;33(3-4):188-201. https://doi.org/10.1089/hum.2021.089
- Devaraju N, V Rajendiran, Ravi NS and Mohankumar KM. Genome engineering of hematopoietic stem cells using CRISPR-Cas9 system. Methods in Molecular Biology. 2429:307-331. 2022. https://doi.org/10.1007/978-1-0716-1979-7_20
- Prasad K, George A, Ravi NS, Mohankumar KM. CRISPR/Cas based gene editing: Marking a new era in medical science. Molecular Biology Reports. 48(5):4879-4895. 2021. https://doi.org/10.1007/s11033-021-06479-7
- Kupp R, Ruff L, Terranova S, Nathan E, Ballereau S, Stark R, Sekhar Reddy Chilamakuri C, Hoffmann N, Wickham-Rahrmann K, Widdess M, Arabzade A, Zhao Y, Varadharajan S, Zheng T, Mohankumar KM, Pfister SM, Kawauchi D, Pajtler KW, Deneen B, Mack SC, Masih KE, Gryder BE, Khan J, Gilbertson RJ. ZFTA-translocations constitute ependymoma chromatin remodeling and transcription factors. Cancer Discovery. 19:1052, 2021. https://doi.org/10.1158/2159-8290.CD-20-1052
- Bagchi A, Nath A, Thamodaran V , Ijee S , Palani D , Rajendiran V , Venkatesan V, Datari P, Pai A , Janet N , Balasubramanian P , Nakamura Y , Srivastava A, Mohankumar KM, Thangavel S and Velayudhan S. Direct Generation of Immortalized Erythroid Progenitor Cell Lines from Peripheral Blood Mononuclear Cells. Cells. 10:523, 2021. https://doi.org/10.3390/cells10030523
- Dharmalingam P, Rachamalla HK, Lohchania B, Bandlamudi B, Thangavel S, Mohankumar KM, Banerjee R, Chaudhuri A, Voshavar C, Marepally S. Green Transfection: Cationic Lipid Nanocarrier System Derivatized from Vegetable Fat, Palm stearin Enhances Nucleic Acid Transfections. ACS Omega 30:7892-7903. 2017. https://doi.org/10.1021/acsomega.7b00935.
- Mohankumar KM, Currle DS, White E, Boulos N, Dapper J,Eden C, Nimmervoll B, Thiruvenkatam R, Connelly M, Kranenburg TA, Neale G, Olsen S, Wang M, Finkelstein D, Wright K, Gupta K, Ellison DW, Thomas AO, Gilbertson RJ. In vivo oncogenomics screen identifies novel ependymoma oncogenes and tumor suppressors within large chromosomal alterations. Nature Genetics. 47:878-87, 2015. https://doi.org/10.1038/ng.3323
- Matthew Parker, Mohankumar KM, Punchihewa C , Weinlich R, Dalton JD, Lee R, Ruth G, Tatevossian RG,. Phoenix TN, Thiruvenkatam R, Li Y, White E, Tang B, Orisme W, Gupta K, Rusch M, Chen X, Nagahawhatte P, Hedlund E, Finkelstein D, Wu G, Shurtleff S, Easton J, Boggs K, Yergeau D, Vadodaria B, Mulder HL, Becksford J, Gupta P, Huether R, Ma J, Song G, Ding L, Lu C, Ochoa K, Zhao D, Fulton RS, Fulton LL, Mardis ER, Wilson RK, Downing JR, Green D, Zhang J, Ellison DW, and Gilbertson RJ. Highly recurrent C11ORF95 fusions drive oncogenic NF-?B signaling in Ependymoma. Nature. 500:451-455, 2014. https://doi.org/10.1038/nature13109
- Johnson RA, Wright KD, Poppleton H, Mohankumar KM, Finkelstein D, Pounds SB, Rand V, Leary SE, White E, Eden C, Hogg T, Northcott P, Mack S, Neale G, Wang YD, Coyle B, Atkinson J, DeWire M, Kranenburg TA, Gillespie Y, Allen JC, Merchant T, Hoop FA, Sanford RA, Gajjar A, Ellison DW, Taylor MD, Grundy RG, Gilbertson RJ. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 466:632-6, 2010. https://doi.org/10.1038/nature09173
- Atkinson JM, Shelat AA, Carcaboso AM, Kranenburg TA, Arnold LA, Boulos N, Wright K, Johnson RA, Poppleton H, Mohankumar KM, Féau C, Phoenix T, Gibson P, Zhu L, Tong Y, Eden C, Ellison DW, Priebe W, Koul D, Yung WK, Gajjar A, Stewart CF, Guy RK, Gilbertson RJ. An integrated in vitro and in vivo high-throughput screen identifies treatment leads for ependymoma. Cancer Cell. 20:384-99, 2011. https://doi.org/10.1016/j.ccr.2011.08.013
- Mohankumar KM, Xu XQ, Zhu T, Kannan N, Miller L, Liu ET, Gluckman PD, Sukumar S, Emerald BS, Lobie PE. HOXA1-stimulated oncogenicity is mediated by selective upregulation of components of the p44/42 MAP kinase pathway in human mammary carcinoma cells. Oncogene. 26: 3998-4008, 2007. https://doi.org/10.1038/sj.onc.1210180
- Eden CJ, Ju B, Mohankumar KM, Phoenix T, Nimmervoll B, Tong Y, Ellison DW, Lessman C, Taylor MR, Gilbertson RJ. Orthotopic models of pediatric brain tumors in zebrafish. Oncogene. 2015 Mar 26;34(13):1736-42, 2014. https://doi.org/10.1038/onc.2014.107
- Mohankumar KM, Perry JK, Kannan N, Kohno K, Gluckman PD, Emerald BS, Lobie PE. Transcriptional activation of STAT3 and STAT5B is important for HOXA1 mediated oncogenic transformation of immortalized human mammary epithelial cells. Endocrinology.149: 2219-29, 2008. https://doi.org/10.1210/en.2007-1320
- Evseenko DA, Murthi P, Paxton JW, Reid G, Emerald BS, Mohankumar KM, Lobie PE, Brennecke SP, Kalionis B, Keelan JA. The ABC transporter BCRP/ABCG2 is a Placental survival factor and its expression is reduced in idiopathic human fetal growth restriction. FASEB J. 21:3592-605, 2007. https://faseb.onlinelibrary.wiley.com/doi/epdf/10.1096/fj.07-8688com
- Pandey V, Perry JK, Mohankumar KM, Kong XJ, Liu SM, Wu ZS, Mitchell MD, Zhu T, Lobie PE. Autocrine human growth hormone stimulates oncogenicity of endometrial carcinoma cells. Endocrinology.149:3909-19, 2008. https://doi.org/10.1210/en.2008-0286
- Zhang X, Emerald BS, Mukhina S, Mohankumar KM, Kraemer A, Yap AS, Gluckman PD, Lee KO, Lobie PE. HOXA1 is required for E-cadherin dependent anchorage independent survival of human mammary carcinoma cells. J Biol Chem. 281: 6471-81, 2006. https://doi.org/10.1074/jbc.M512666200
- Ramakrishna L, Anand KK, Mohankumar KM, Ranga U. Codon optimization of the Tat antigen of human immunodeficiency virus type 1 generates strong immune responses in mice following genetic immunization. J Virol. 78: 9174-89, 2004. https://doi.org/10.1128/jvi.78.17.9174-9189.2004
- Hou L, Xu B, Mohankumar KM, Goffin V, Perry JK, Lobie PE, Liu DX. The prolactin receptor mediates HOXA1- stimulated oncogenicity of mammary carcinoma cells. Int J Oncol. 41:2285-95, 2012. https://doi.org/10.3892/ijo.2012.1660
- Kannan N, Kang J, Kong X, Tang J, Perry JK, Mohankumar KM, Miller LD, Liu ET, Mertani HC, Zhu T, Grandison PM, Liu DX, Lobie PE. Trefoil factor 3 is Oncogenic and mediates anti-estrogen resistance in human mammary carcinoma. Neoplasia.12:1041-53, 2010. https://doi.org/10.1593/neo.10916
- Ramakrishna L, Anand K, Mahalingam M, Mohankumar KM, Ramani S, Siddappa NB, Ranga U. Codon optimization and ubiquitin conjugation of HIV-1 Tat lead to enhanced cell mediated immune responses. Vaccine. 22: 2586-98, 2004. https://doi.org/10.1016/j.vaccine.2003.12.007
- Mohankumar KM, Bobby Z, Selvaraj N, Das A, Koner BC, Sen SK, Ramesh R, Ranganathan P. Possible link between glycated hemoglobin and lipid peroxidation in hyperthyroidism. Clinical Chemistry Acta. 342: 187-192, 2004. https://doi.org/10.1016/j.cccn.2003.12.027
- Perry JK, Mohankumar KM, Emerald BS, Mertani HC, Lobie PE. The contribution of growth hormone to mammary neoplasia. J Mammary Gland Biol Neoplasia.13:131- 45, 2008. (Review). https://doi.org/10.1007/s10911-008-9070-z
- Chandraprabha, P. B., Azhagiri, M. K. K., Venkatesan, V., Magis, W., Prasad, K., Suresh, S., Pai, A. A., Marepally, S., Srivastava, A., Mohankumar, KM, Martin, D. I. K., & Thangavel, S. (2024). Enhanced fetal hemoglobin production via dual-beneficial mutation editing of the HBG promoter in hematopoietic stem and progenitor cells for β-hemoglobinopathies. Stem cell research & therapy. 10.1186/s13287-024-04117-
- stems and Methods for Gene Modifications in the First Intron of Human Gamma Globin and Fetal Hemoglobin Induction · Application No: 202341082821
- Strategies for Precision Editing of Homologous Regions · Application No: 202341082277
- Enhanced Gene Reactivation via Targeted Promoter Editing · Application No:202441072108
- Therapeutic Rescue of Neutrophil Maturation Arrest by Base Editing of ELANE in Severe Congenital Neutropenia Invention Disclosure Form Submitte
- COMPOSITIONS AND METHODS FOR REACTIVATING DEVELOPMENTALLY SILENT GENES
Application number: 202041020165 - A METHOD FOR MODIFICATION OF ß-GLOBIN GENE
Application number:202241030885 - A METHOD FOR ELEVATION OF GAMMA GLOBIN
Application No: 202241055876 - STRATEGIES FOR PRECISION EDITING OF HOMOLOGUS REGIONS.
Application No: 202341082277 - A METHOD FOR GENE MODIFICATION WITHIN THE INTRON 1 REGION OF GAMMA GLOBIN.
Application No: 202341082821 - ENHANCED GENE REACTIVATION VIA TARGETED PROMOTER EDITING.
Application No: 202441072108 - THERAPEUTIC RESCUE OF NEUTROPHIL MATURATION ARREST BY BASE EDITING OF ELANE IN SEVERE CONGENITAL NEUTROPENIA.
Application No: 202541053473
CSCR/CMC
- Dr Alok Srivastava, CSCR
- Dr R.V. Shaji, CSCR
- Dr Saravanbhavan Thangavel CSCR
- Dr Srujan Marepally, CSCR
- Dr Sanjay Kumar, CSCR
- Dr Poonkuzhali Balasubramanian, Department of Hematology, CMC
- Dr Sukesh Chandran Nair, Department of Transfusion Medicine, CMC
- Dr Premila Abraham and Dr. Muthuraman N, Department of Biochemistry, CMC
- Dr Christhunesa Soundararajan, Department of Neurochemistry, CMC
- Dr. Fouzia N.A, Department of Hematology, CMC.
External
- Dr Merlin Crossley, Deputy Vice-Chancellor, University of New South Wales, Australia
- Dr Jacob Corn, ETH Zürich, Switzerland


















